The underlying core concept of most, if not all, of the high-throughput technology is “coding” and “decoding”. The beauty of phage display technology lies in the super high diversity that it has even for a single library of phage-displayed peptides, proteins or antibodies, and also the physical linkage of the surface presenting peptide/protein to its coding DNA sequence. Thus, we think phage display is the ideal platform for coding. Meanwhile, the Next Generation Sequencing technology possesses the power of highly efficient parallel sequencing, in another word “decoding”. In order to exemplify the “coding” power of phage display and the “decoding” power of NGS, we combined these technologies together, developed an integrated technology, namely, AbMap and used for a variety of applications, e. g., antibody epitope mapping, and disease biomarker discovery.
Antibody epitope mapping
Antibodies play essential roles in both diagnostics and therapeutics. Epitope mapping is essential to understand how an antibody works and to protect intellectual property. Given the millions of antibodies for which epitope information is lacking, there is a need for high-throughput epitope mapping. To address this, we developed a strategy, Antibody binding epitope Mapping (AbMap), by combining a phage displayed peptide library with next generation sequencing. Using AbMap, profiles of the peptides bound by 202 antibodies were determined in a single test, and linear epitopes were identified for >50% of the antibodies. Using spike protein (S1 and S2)-enriched antibodies from the convalescent serum of one COVID-19 patient as the input, both linear and potentially conformational epitopes of spike protein specific antibodies were identified. We defined peptide-binding profile of an antibody as the Binding Capacity (BiC). Conceptually, the BiC could serve as a systematic and functional descriptor of any antibody. Requiring at least one order of magnitude less time and money to map linear epitopes than traditional technologies, AbMap allows for high-throughput epitope mapping and creates many possibilities. Taking advantage of this pipeline, we have already determined the epitopes of 100,000+ antibodies.
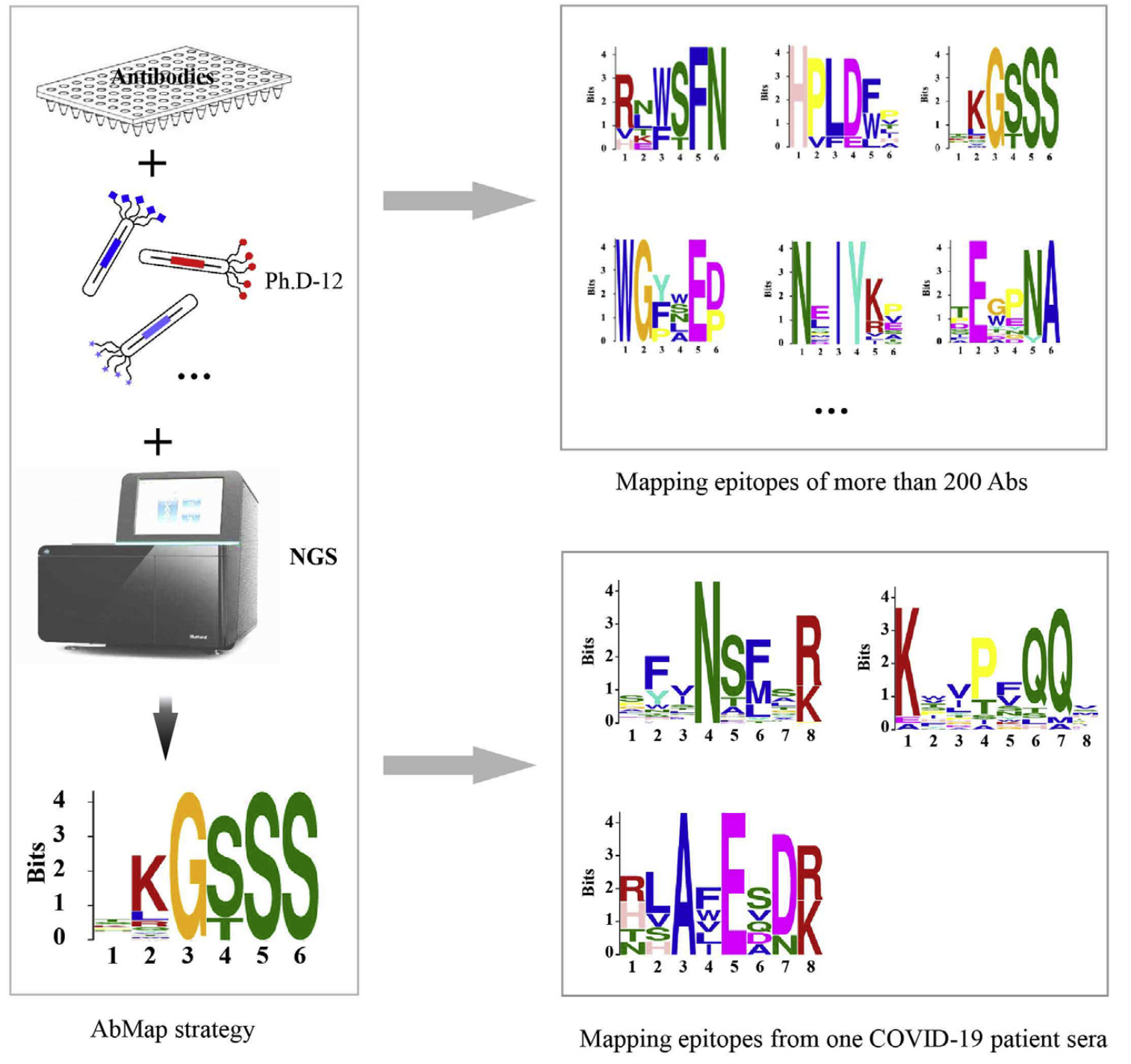
Mapping the antibody recognizing epitope on antigen by AbMap
Disease biomarker discovery
Disease biomarker is always needed for accurate diagnostics and prognostics. However, technology suitable for identifying reliable biomarkers from a large set of samples in a very short period of time is still lacking. In one example, we applied the AbMap technology for identifying Systemic lupus erythematosus (SLE) serum biomarkers. SLE is one of the most serious autoimmune diseases, characterized by highly diverse clinical manifestations. Biomarker is still needed for the accurate diagnostics. SLE serum autoantibodies were discovered and validated using serum samples from independent sample cohorts encompassing 306 participants divided into three groups, i.e., healthy, SLE patients, and other auto-immune related diseases. To discover biomarkers for SLE, a phage displayed random peptide library (Ph.D. 12) and deep sequencing were applied to screen specific autoantibodies in a total of 100 serum samples from 50 SLE patients and 50 healthy controls. Ten peptides were validated using enzyme linked immunosorbent assays (ELISA). As a result, four peptides were discovered with high diagnostic power to differentiate SLE patients from health controls. Among them, two peptides were confirmed between SLE with other auto-immune patients. The procedure we established could be easily adopted for the identification of autoantibodies as biomarkers for many other diseases.
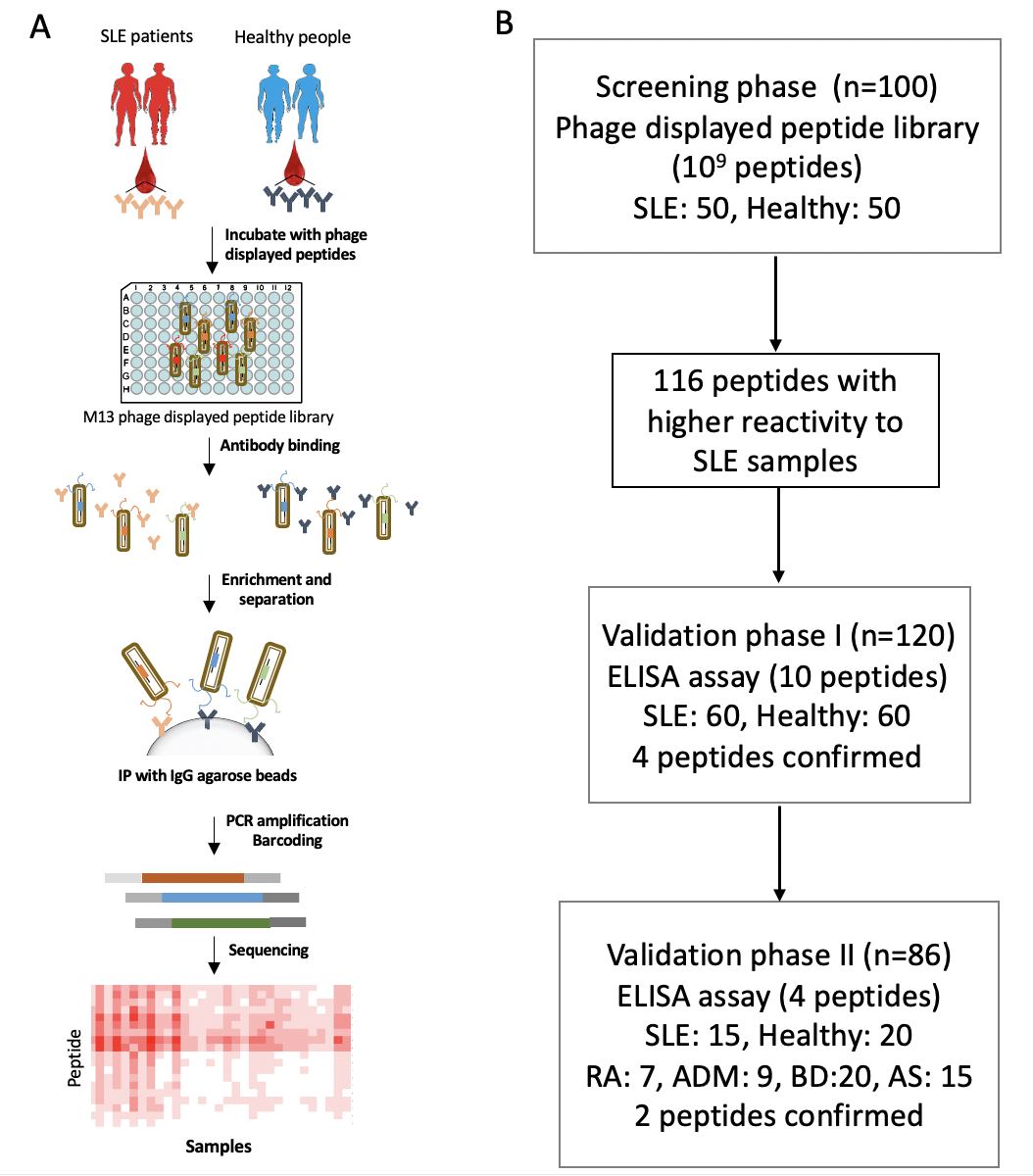
Procedure for SLE serum biomarker discovery by using AbMap